Phylogenetic Biology - KnowledgeVis is the software lead for the Arbor Workflows project. Arbor’s PhyloMap interface allows phylogenetic biologists to explore a tree of related organisms through their place in the tree or through the geospatial location of instances.
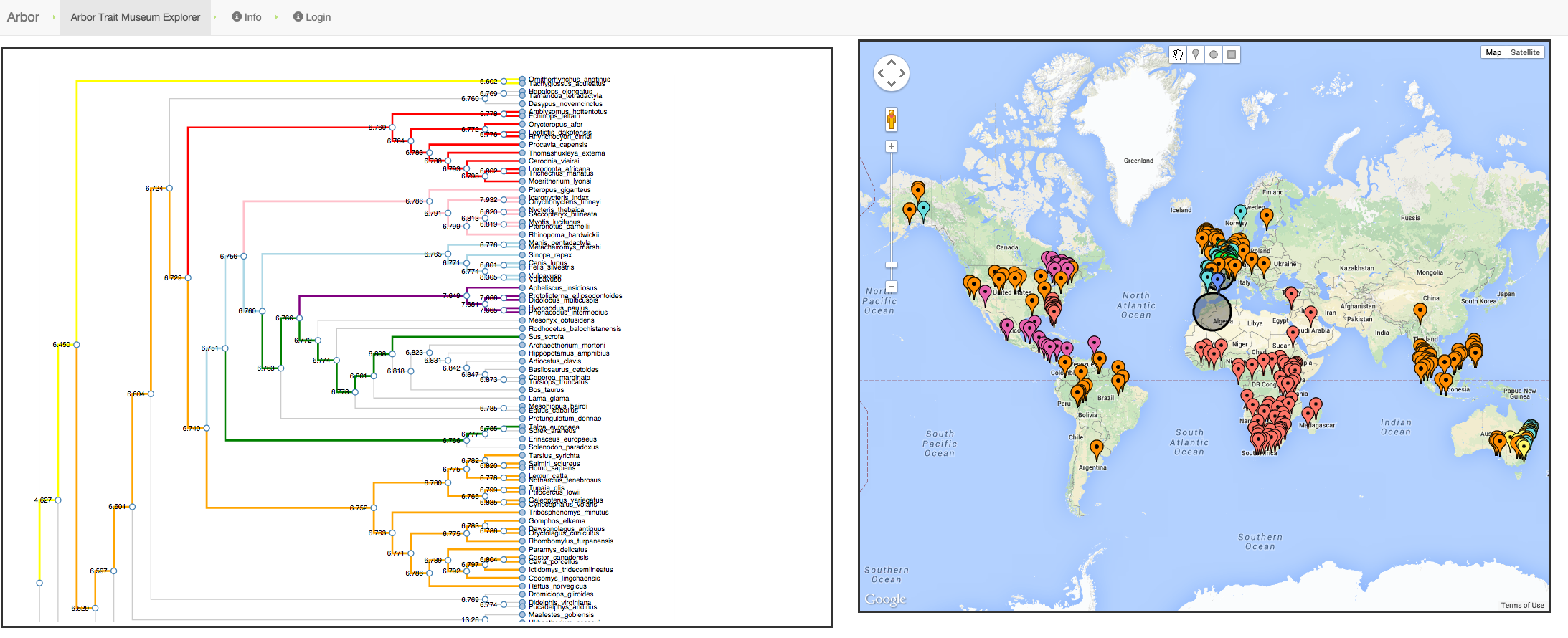
Exploring the distribution of species with Arbor
Medical Image Processing - CT and MR
clinical imaging systems, along with confocal microscopy devices,
generate volumetric datasets that lend themselves to automated
processing by computer algorithms. As these datasets increase
in size, parallel processing and multithreading algorithms are
of greater necessity. KnowledgeVis has experience innovating with
deep learning tools, and open source products, such as the
Visualization Toolkit (VTK) and the Insight Toolkit (ITK)
to analyze and process images from multiple modalities.
Below is a finite element model of a finger bone rendered
by a KnowledgeVis developed rendering algorithm that
extracts bone density values from a source MRI image.
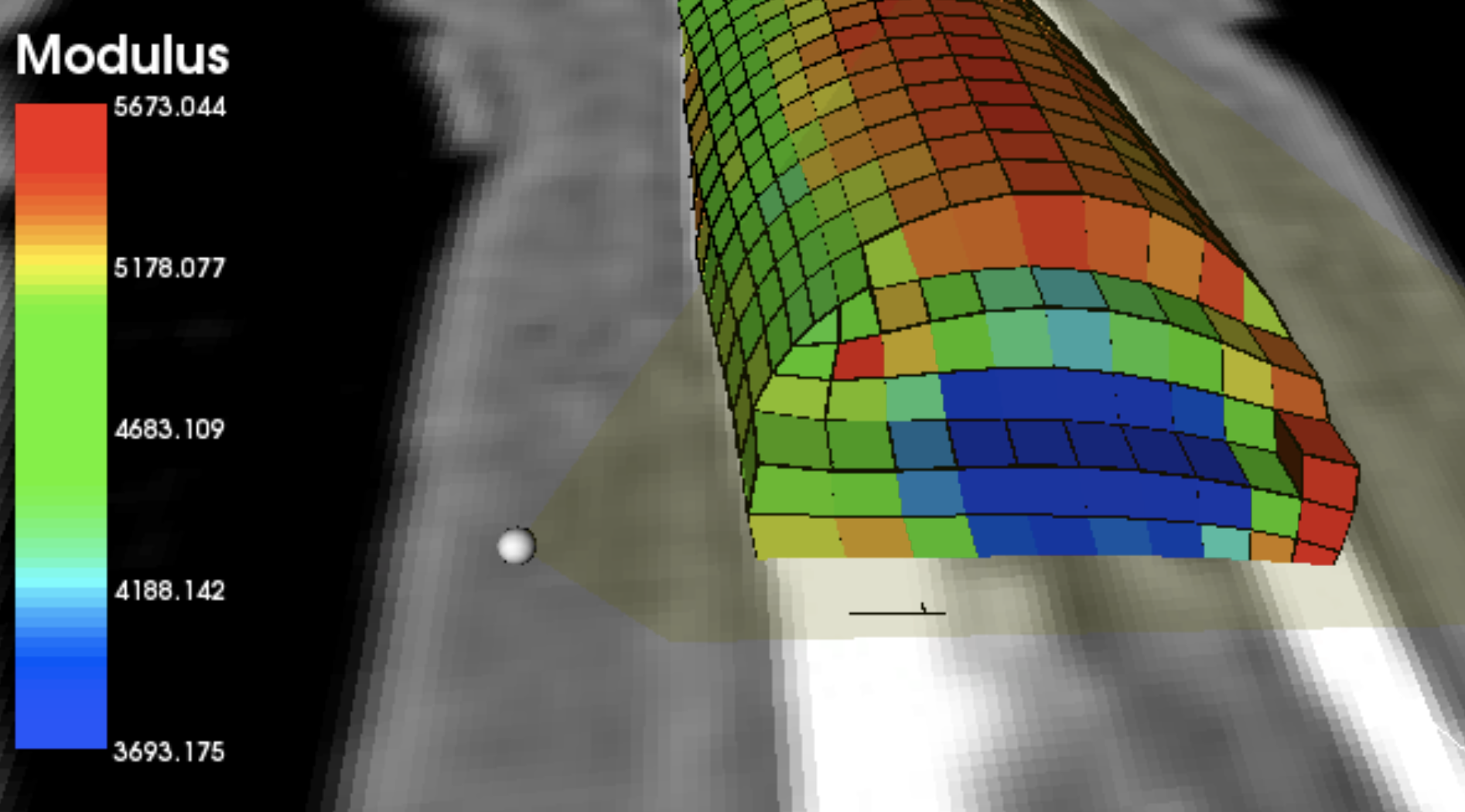
3D Bone Density Visualization
DARPA XDATA - KnowledgeVis developed visualization tools for large network and geotemporal datasets. This project was all about methods to store, retrieve, and analyze very large datasets. These interfaces below queried data from a mongoDB instance and rendered using javascript libraries.
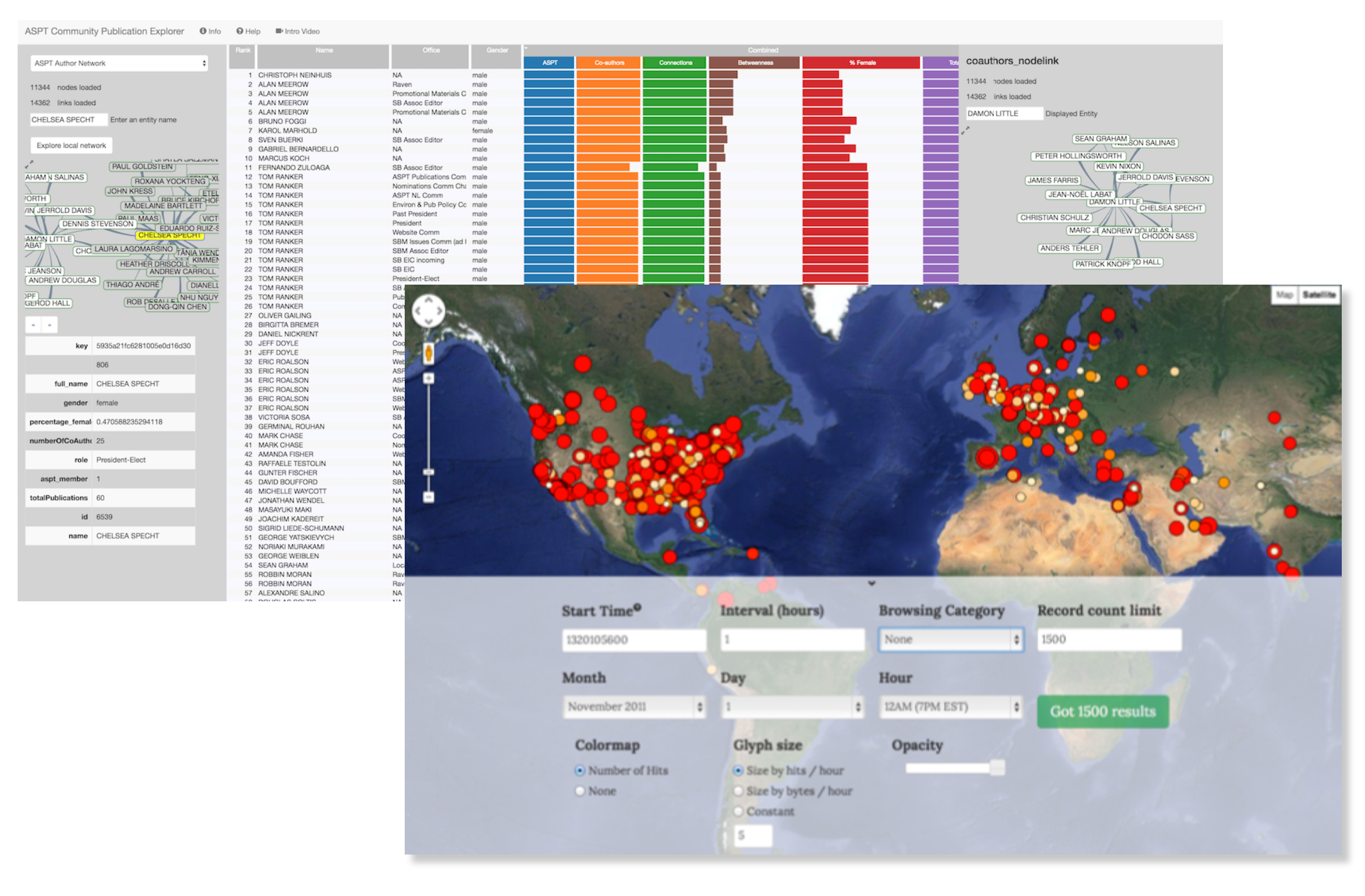
Exploring Network data during XDATA
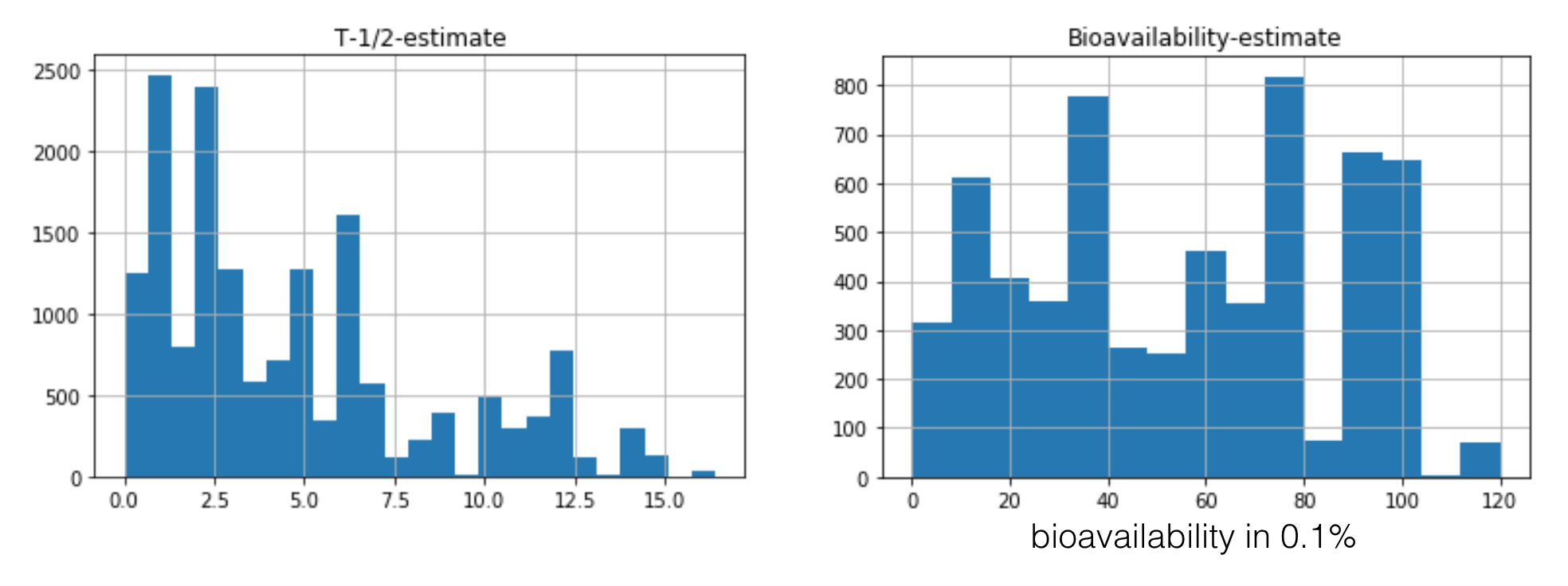
Natural Language Processing - KnowledgeVis developed NLP solutions to process free text in academic journals and drug labels. These solutions automatically extracted feature names and values from the source texts. As an example, half-life and bioavailability histograms are shown for a suite of drug compounds. The values were automatically extracted from drug label discriptions using NLP techniques written using the Python computer language.